Integrated Genomic Surveillance (IGS)
What is integrated genomic surveillance?
Integrated genomic surveillance (IGS) is an effective public health strategy for monitoring infectious pathogens, the infectious diseases that they cause, and transmission events that is increasingly gaining importance worldwide.
A legal basis for molecular/genomic surveillance in Germany was established with the amendment of §13 of the German Infection Protection Act (Infektionsschutzgesetz, IfSG) in July 2017 and the introduction of the Measles Protection Act in March 2020.
IGS involves combining data from different sources. Specifically, the results of modern DNA sequencing methods and genome sequence analysis methods for pathogen isolates from patient samples are merged with epidemiological information from the reporting system according to the IfSG and further pathogen data.
This aggregated data can then be quickly and systematically analysed with regard to different questions.
 Integrated genomic surveillance. Source: © RKI
Integrated genomic surveillance. Source: © RKI
About the terms IMS and IGS
The term “integrated molecular surveillance (IMS)” was previously coined at the RKI for a project involving the aggregation of epidemiological and molecular genetic information with regard to the monitoring of infectious diseases. In an international context, and especially since the COVID-19 pandemic, the term “genomic surveillance” is mainly used. In alignment with this, the term “integrated genomic surveillance (IGS)” will henceforth be used when the focus is on genomic data. The term “integration” refers to the amalgamation and joint examination of epidemiological reporting data, genomic data, and further molecular pathogen data. The term “molecular surveillance”, as used in IfSG (§13), for example, encompasses genomic surveillance as well as other molecular characteristics of a pathogen that may be important for the differentiation of pathogens.
What objectives are to be achieved with integrated genomic surveillance?
The main objective of integrated genomic surveillance is the timely and nationwide detection of a noticeable spread of infectious pathogens and the identification of associations between identified pathogens even when they are temporally and spatially separated. IGS can reveal relationships that would otherwise remain undiscovered. Sequence comparisons thus uncover possible genetic similarities between identified pathogens, which can indicate associations such as common sources or undetected contacts.
In combination with selected information about the people affected by the infection, outbreaks and transmission chains can be identified more quickly and targeted interventions can be initiated at an early stage to contain the outbreak. IGS also enables the continuous monitoring of the transmission event, even beyond unusual occurrences.
Besides the recognition of transmission associations, IGS offers the possibility of the continuous identification and monitoring of pathogenic variants with changed properties (e.g. variants with changed transmissibility, resistance factors to antimicrobial therapeutics, or changed virulence). This knowledge can be used to respond to changes in the properties of pathogens or their transmission.
In addition, IGS makes an important contribution to the decision-making processes of health authorities. Using IGS, action-oriented information is provided on the basis of an overall view of the regional distribution of pathogenic variants combined with epidemiological information from the reporting system. Furthermore, IGS ensures that the RKI, the German public health service (ÖGD), and the German Federal Ministry of Health (BMG) are able to provide national stakeholders and international institutions, such as the ECDC and the WHO, with information about infection events. In this way, IGS contributes significantly to the optimisation of measures to prevent cases of disease and deaths.
How is integrated genomic surveillance implemented?
If a notifiable case according to §7.1 IfSG involving a pathogen that is monitored within the framework of IGS is detected, multiple steps are carried out in parallel:
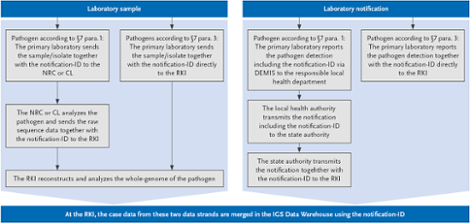
Scheme of the IGS at the RKI. Source: © RKI
Meldungs-ID: Identification number, which must be generated by the detector according to a defined scheme
DEMIS: German Electronic Reporting and Information System for Infection Protection
IGS-Data-Warehouse: Data Management System
What aspects are important for successful integrated genomic surveillance?
- Clinical awareness of the occurrence of infectious diseases.
- The initiation of the necessary pathogen diagnostics at the consulted medical practice.
- The proper extraction and processing of isolates.
- A defined sample and data flow (clinic/medical practice > primary diagnosing laboratory > regional laboratory/national reference centre (NRC)/consultant laboratory (CL) > RKI).
- The storage and merging of relevant pathogen data with epidemiological data
- The central epidemiological analysis of the combined data.
- The reporting back of results to the ÖGD and other stakeholders to initiate possible countermeasures.
The short film below shows how IGS works and its potential benefits using the example of antibiotic-resistant pathogens:
 Integrated genomic surveillance in Germany using the example of antibiotic-resistant pathogens. Source: © RKI
Integrated genomic surveillance in Germany using the example of antibiotic-resistant pathogens. Source: © RKI
Which stakeholders are involved in IGS?
In order to amalgamate data from different sources and enable subsequent evaluations, close cooperation between health authorities, medical microbiological laboratories, and public health facilities is necessary. Additionally, appropriate data infrastructure must be available and data exchange must be ensured. In the context of IGS, this is implemented via the German electronic reporting and information system DEMIS and SurvNet.
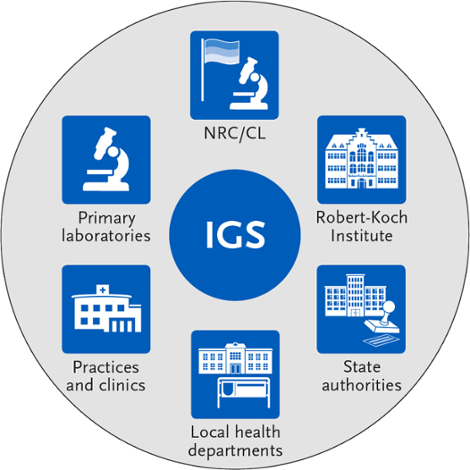 Stakeholders involved in the IGS. Source: © RKI
Stakeholders involved in the IGS. Source: © RKI
Integrated genomic surveillance at the RKI
It is urgently necessary to also develop strong infrastructure in Germany in order to perform systemic integrated genomic surveillance at the national level.
The RKI began establishing IGS back in 2014 in the form of modules and pilot studies. In the first step, the initial financing provided by special research funds of the RKI and projects of the Federal Ministry of Health focused on food-borne pathogens (EHEC, listeria, and salmonella) as well tuberculosis (TB) and HIV.
Based on the significant structural and substantive results of these projects, the performance of IGS for SARS-CoV-2 began in the context of the pandemic event. Since 2021 and 2022, an internationally comparable IGS funded by the Federal Ministry of Health is being developed in Germany (project duration until the end of 2025). Its objective is to design, establish, and evaluate the components of an IGS at the RKI with the involvement of the national reference centres (NRC) and consultant laboratories (CL). The processes required for this are to be established and effective infrastructure for the sustainable expansion of IGS according to §13 of the IfSG is to be developed. The developed processes are to be made permanent beyond the end of the project.
The prioritisation of pathogens and infectious diseases subject to IGS is periodically carried out by the RKI on the basis of the respective present situation and international recommendations, in particular from the ECDC and the WHO. Various epidemiological, microbiological, and legal aspects are taken into account and a three-level prioritisation system (A - high, B - medium, C - low) is used.
Overview of pathogens relevant for the design of IGS at the RKI by priority of the intended implementation. The pathogens in the table are first sorted by priority of implementation (A - high, B - medium, C - low) and then alphabetically within these categories.
| Pathogens | Transmission routes | Recommended survey mode | Priority of implementation |
|---|
| EHEC / HUS | FBI | Full survey | A |
| Listeria monocytogenes | FBI | Full survey | A |
| Salmonella Enteritidis | FBI | Full survey | A |
| Salmonella Typhimurium | FBI | Full survey | A |
| (Human pathogenic) influenza viruses | RESP | Partial survey | A |
| Invasive Neisseria meningitidis | RESP | Full survey | A |
| Measles virus | RESP | Full survey | A |
| Mycobacterium tuberculosis | RESP | Full survey | A |
| SARS-CoV-2 | RESP | Partial survey | A |
| HIV | STI | Full survey | A |
| Less sensitive Neisseria gonorrhoeae | STI | Partial survey | A |
| Carbapenem-resistant Enterobacterales | NI | Full survey | A |
| Carbapenem-resistant Acinetobacter baumannii | NI | Full survey | B |
| Clostridioides difficile | NI | Partial survey | B |
| Methicillin-resistant Staphylococcus aureus (MRSA) | NI | Partial survey | B |
| Salmonella, other serovars | FBI | Event-related | B |
| Hepatitis E virus | FBI | Partial survey | C |
| Hepatitis A virus | FBI/STI | Full survey | C |
| Legionella | RESP | Event-related | C |
| Pneumococci | RESP | Full survey | C |
| West Nile virus | VB | Full survey | C |
Notes: The transmission routes were divided into four categories. The objective of a full survey of a pathogen is to examine all cases that can be technically typified.
Acronyms:
| FBI | Food-borne infections |
| RESP | Respiratory infections |
| NI | Nosocomial infections |
| STI | Sexually transmitted infections |
| VB | Vector-borne infections |
What added value does IGS bring with regard to respiratory pathogens such as SARS-CoV-2?
In the case of respiratory pathogens such as SARS-CoV-2, timely IGS can make an important contribution to the assessment of an epidemic event as it continuously provides current information on the pathogens in the context of their spread (e.g. virus variants with new characteristics) and thus allows for a valid appraisal of the epidemiological situation. Furthermore, sound health policy decisions can be made on the basis of the IGS data. The general public can also be fully informed. Scientists are able to determine the burden of disease within the population and adapt diagnostic tests, treatments, and vaccines to the respective current situation.
How can IGS be beneficial with regard to food-borne infections such as Salmonella Braenderup?
Even with geographically widespread distribution and without a local disease cluster, IGS enables the detection of closely related pathogens and thus the identification of an outbreak with common sources. An example of this is the emergence of Salmonella Braenderup in 13 countries (likely caused by contaminated Galia melons), which was determined to be an outbreak.
What added value does IGS bring with regard to nosocomial infections and antibiotic-resistant pathogens?
Antibiotic-resistant pathogens can cause infections that are difficult to treat and, in particular, play an important role in the nosocomial setting. Nationwide surveillance of widespread species, their strains, and resistance factors is therefore important for public health measures and clinical decisions.
Local outbreaks of antibiotic-resistant pathogens, e.g. in a hospital, are usually identified and combated on site. Supra-regional or international outbreaks can occur as a result of undetected infection chains, e.g. due to patient transfers or contaminated food or medical devices. With a supra-regional detection of outbreaks based on isolates collected nationwide and reporting information within the framework of IGS, outbreaks can be detected more quickly and transmission paths (e.g. horizontal vs. clonal transmission) can be understood more quickly, thus enabling more effective public health measures to be taken.
The general distribution of nosocomial, antibiotic-resistant pathogens and their resistance factors are also to be tracked as part of IGS. Information about regionally prevalent pathogens and resistance mechanisms can influence clinical therapeutic decisions.
What added value does IGS bring with regard to sexually transmitted and blood-borne pathogens?
Monitoring primary HIV resistance and HIV diversity is one of the official tasks of the Robert Koch Institute. For this purpose, resistance-relevant HIV genome areas have been sequenced since 2013 from a large proportion of newly diagnosed HIV patients who are not reported by name to the RKI, and a genotypic HIV resistance determination has been carried out. In addition, the type and subtype and, if appropriate, the recombinant form are determined from the HIV sequences. In more extensive analyses, HIV transmission clusters are identified and the German HIV epidemic is characterized phylodynamically and phylogeographically on the basis of epidemiological data. As part of the integrated genomic surveillance of HIV (IGS-HIV), near real-time surveillance using laboratory and bioinformatic automation is currently being set up. This will make it possible to identify and present the dynamics of HIV outbreaks even more quickly and more comprehensively. Another important aspect of the HIV epidemic is the varying length of time between HIV infection and HIV diagnosis. Therefore, a bioinformatic method is to be developed that can be used to estimate the duration of a newly diagnosed HIV infection based on HIV-NGS data. In combination with epidemiological data, risk groups with delayed HIV diagnoses or an acutely high incidence of infection can thus be identified. In summary, IGS-HIV provides important information for the further development of testing and prevention strategies as well as diagnostic and treatment guidelines. Detailed information about the surveys and projects can be found on the pages of the HIV study laboratory.
Another important focus of integrated genomic surveillance at the RKI is the study of the resistance developments of STIs. The anti-microbial resistance (AMR) of gonococci (Neisseria gonorrhoeae), the pathogens causing gonorrhoea, is a growing problem worldwide. In recent decades, the pathogen has developed resistances against all main classes of antibiotics. The pharynx is an important reservoir for gonococci and their genesis of resistance. However, because pharyngeal infections are usually asymptomatic and culturing samples from the pharynx is often difficult, little data is available on AMR in this location. For this reason, isolated gonococci, especially from the pharynx, are to be systematically phenotypically and genotypically examined with regard to AMR as part of integrated genomic surveillance. Detailed molecular data allow for a phylogenetic characterisation of the isolates. Together with further data from national and international genomic data, these data can be used to map larger transmission networks and analyse the dynamics of the spread of AMR within these networks. On this basis, it is possible to derive necessary adjustments to the treatment guidelines and the antibiotics recommended therein, and resistance-guided therapy can be implemented, thus sparing the patient from ineffective and excessively frequent treatment with antibiotics.
Current IGS projects at the RKI
Completed IGS projects
Further IGS-relevant activities
Contact information
We are available for information, comments and suggestions at the e-mail address IGS@rki.de. State authorities and health authorities should contact surveillance@rki.de.
to the top